论文标题:Multiple domains of bacterial and human Lon proteases define substrate selectivity
期刊:Emerging Microbes & Infections
作者:Lihong He et al
发表时间: 2018/8/17
数字识别码:10.1186/s41426-018-0148-4
原文链接:https://www.nature.com/articles/s41426-018-0148-4?utm_source=Other_website&utm_medium=Website_linksWebsite_links&utm_content=JesGuo-Nature-Emerging_Microbes_and_Infections-Biology-China&utm_campaign=NROAAJ_USG_JRCN_JG_NROAAJ_Multiple
Lon 是ATP酶超家族的一员,在原核生物和真核生物中发挥着许多重要的功能。在细菌里, Lon被认为是将错误折叠蛋白降解的主要蛋白酶,它依靠蛋白水解来调节许多重要功能,包括封装、遗传能力、运动性、热休克反应、持久性和耐药性、 DNA的复制和修复以及毒力因子的产生。人Lon位于线粒体基质,它的功能包括对线粒体里的蛋白质的质量控制,维持线粒体DNA类核的完整性,以及对缺氧和氧化应激的反应。Lon的功能障碍和许多疾病相关,包括衰老、癌症,和脑-眼-牙-耳-骨骼综合症有关。然而,我们对Lon如何选择蛋白质底物来降解的分子机制, 以及Lon蛋白酶在原核生物和真核生物里的有哪些变体了解不多。
在《新发现病原体与感染》杂志里一篇名为“Multiple domains of bacterial and human Lon proteases define substrate selectivity”的文章里,清华大学医学院传染病研究中心张敬仁教授带领的团队和四川大学华西医院生物治疗国家重点实验室合作,研究了与细菌和人Lon选择底物有关的结构域。
在 21 个候选底物里,作者们识别其中10 个为土拉弗菌的底物。这是有史以来识别最多 Lon 底物的一次研究。作者们发现,人Lon和细菌Lon之间没有跨物种的活性,即人Lon不能降解细菌Lon的底物,所以Lon对底物的选择可能是因物种而异。如果把人Lon的N、A, 或P结构域逐一换成细菌Lon里的相对的结构域,人Lon还是不能降解细菌Lon的底物。这显示,因物种而异的Lon对底物的选择同时受多个结构域的影响。通过体外蛋白水解和质谱分析,研究者发现细菌和人Lon分裂底物的模式相似,这显示Lon在选择同物种的底物时对结构上的契合是有高度的要求的。
研究者认为,这里的研究结果在未来可能会推进蛋白酶或蛋白酶工程的重新设计。
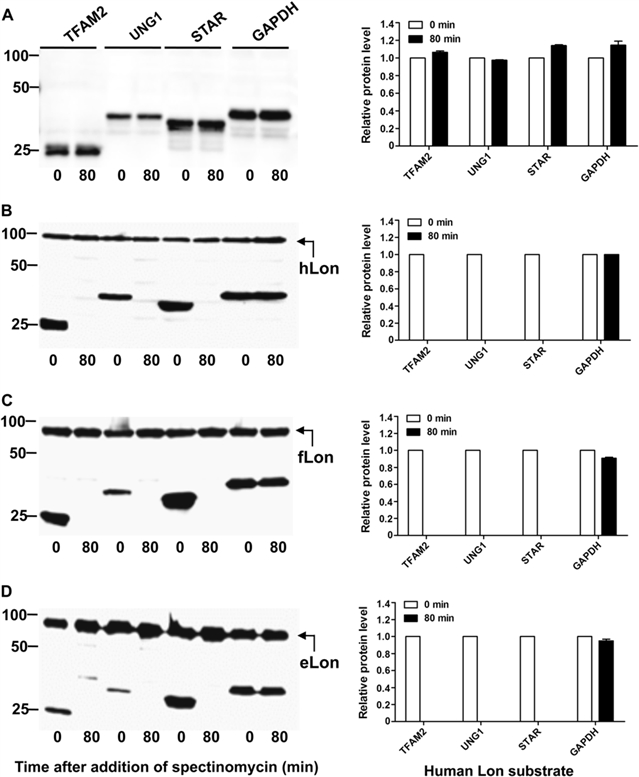
图:人Lon(hLon)对细菌Lon的底物的跨物种的降解。
Abstract:
The Lon protease selectively degrades abnormal proteins or certain normal proteins in response to environmental and cellular conditions in many prokaryotic and eukaryotic organisms. However, the mechanism(s) behind the substrate selection of normal proteins remains largely unknown. In this study, we identified 10 new substrates of F. tularensis Lon from a total of 21 candidate substrates identified in our previous work, the largest number of novel Lon substrates from a single study. Cross-species degradation of these and other known Lon substrates revealed that human Lon is unable to degrade many bacterial Lon substrates, suggestive of a “organism-adapted” substrate selection mechanism for the natural Lon variants. However, individually replacing the N, A, and P domains of human Lon with the counterparts of bacterial Lon did not enable the human protease to degrade the same bacterial Lon substrates. This result showed that the “organism-adapted” substrate selection depends on multiple domains of the Lon proteases. Further in vitro proteolysis and mass spectrometry analysis revealed a similar substrate cleavage pattern between the bacterial and human Lon variants, which was exemplified by predominant representation of leucine, alanine, and other hydrophobic amino acids at the P(−1) site within the substrates. These observations suggest that the Lon proteases select their substrates at least in part by fine structural matching with the proteins in the same organisms.
阅读论文原文,请访问
https://www.nature.com/articles/s41426-018-0148-4?utm_source=Other_website&utm_medium=Website_linksWebsite_links&utm_content=JesGuo-Nature-Emerging_Microbes_and_Infections-Biology-China&utm_campaign=NROAAJ_USG_JRCN_JG_NROAAJ_Multiple
(来源:科学网)
特别声明:本文转载仅仅是出于传播信息的需要,并不意味着代表本网站观点或证实其内容的真实性;如其他媒体、网站或个人从本网站转载使用,须保留本网站注明的“来源”,并自负版权等法律责任;作者如果不希望被转载或者联系转载稿费等事宜,请与我们接洽。