论文题目:Genome sequencing of the staple food crop white Guinea yam enables the development of a molecular marker for sex determination
期刊:BMC Biology
作者:Muluneh Tamiru et al.
发表时间:2017/9/19
数字识别码:10.1186/s12915-017-0419-x
原文链接:https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-017-0419-x?utm_source=WeChat&utm_medium=Social_media_organic&utm_content=DaiDen-BMC-BMC_Biology-Biology-China&utm_campaign=BMCF_USG_Article
“环境与可持续性”主题集精选了BMC Biology发表的关于人类世中生态学、可持续性发展和环境挑战等方面的研究论文、评论、综述和问答文章,精选此篇,供大家阅览!
在BMC Biology发表的研究Genome sequencing of the staple food crop white Guinea yam enables the development of a molecular marker for sex determinatio中,日本研究岩手生物技术研究中心 (Iwate Biotechnology Research Center,IBRC) Ryohei Terauchi领导的团队破译了几内亚山药的基因组。
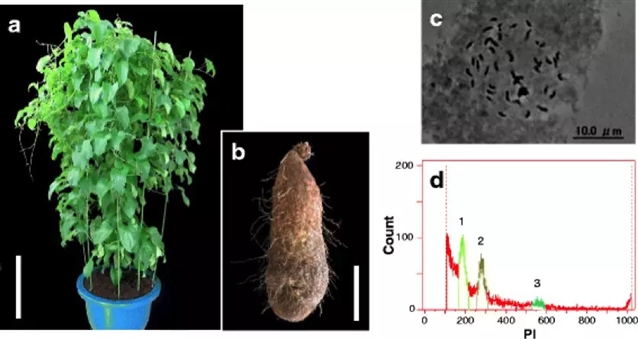
根茎类作物是热带非洲的主要食物来源,其中包括单子叶植物中的薯蓣属(Dioscorea)下的几个品种,俗称山药。山药是上成百上千万人赖以生存的主食,也是社会文化生活的重要组成部分,在西非和中非更是如此。山药种植受许多因素限制,一些改善作物的研究本可以提升他们的种植和品质,然而山药几乎可以被看作作物中被忽视的“孤儿”,缺乏基因和基因组工具阻碍了这种主粮作物的优化进程。
实验结果
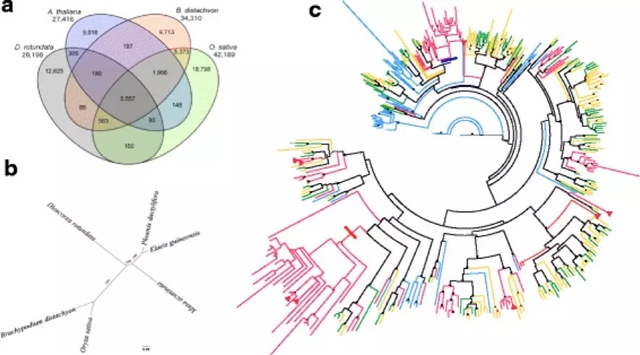
为了加速有标记辅助的山药育种,作者及其研究团队对于几内亚白山药(Dioscorea rotundata)进行了基因组分析,获得了594兆的基因组。其中76.4%分布在21个联锁群上。他们预测白山药共有26,198个基因。对2381个保守基因进行系统发育分析发现,薯蓣属(Dioscorea )是单子叶植物下的一个独特品系,与禾本目(水稻)、棕榈目(棕榈)、姜目(香蕉)均不相同。整个薯蓣属的典型特征是雌雄异株(dioecy),这一特征限制了山药的育种效率。为了推断性别决定的遗传机理,作者对F1子代分离雌株和雄株进行了全基因组混合集群的重测序(数量性状位点测序,QTL-seq),识别出了一个与雌性异形配子(雄性基因型ZZ,雌性基因型ZW)性别决定相关的基因组区域。研究人员们进一步确定了W位点,并利用它研究出了一个能够在幼苗阶段识别出几内亚山药性别的分子标记。
结论
几内亚山药是一类独特且高度分化的单子叶植物进化枝。本次研究中进行的基因组分析和性别关联标记物研发可以极大地加速由标记辅助的几内亚山药育种。除此之外,该QTL-seq分析方法也可以应用于其他异型杂交作物或拥有高度杂合基因组的物种。对像山药这样的孤儿作物进行基因组分析可以提升食物安全,同时推动热带农业的可持续发展。
摘要:
Background
Root and tuber crops are a major food source in tropical Africa. Among these crops are several species in the monocotyledonous genus Dioscorea collectively known as yam, a staple tuber crop that contributes enormously to the subsistence and socio-cultural lives of millions of people, principally in West and Central Africa. Yam cultivation is constrained by several factors, and yam can be considered a neglected “orphan” crop that would benefit from crop improvement efforts. However, the lack of genetic and genomic tools has impeded the improvement of this staple crop.
Results
To accelerate marker-assisted breeding of yam, we performed genome analysis of white Guinea yam (Dioscorea rotundata) and assembled a 594-Mb genome, 76.4% of which was distributed among 21 linkage groups. In total, we predicted 26,198 genes. Phylogenetic analyses with 2381 conserved genes revealed that Dioscorea is a unique lineage of monocotyledons distinct from the Poales (rice), Arecales (palm), and Zingiberales (banana). The entire Dioscorea genus is characterized by the occurrence of separate male and female plants (dioecy), a feature that has limited efficient yam breeding. To infer the genetics of sex determination, we performed whole-genome resequencing of bulked segregants (quantitative trait locus sequencing [QTL-seq]) in F1 progeny segregating for male and female plants and identified a genomic region associated with female heterogametic (male = ZZ, female = ZW) sex determination. We further delineated the W locus and used it to develop a molecular marker for sex identification of Guinea yam plants at the seedling stage.
Conclusions
Guinea yam belongs to a unique and highly differentiated clade of monocotyledons. The genome analyses and sex-linked marker development performed in this study should greatly accelerate marker-assisted breeding of Guinea yam. In addition, our QTL-seq approach can be utilized in genetic studies of other outcrossing crops and organisms with highly heterozygous genomes. Genomic analysis of orphan crops such as yam promotes efforts to improve food security and the sustainability of tropical agriculture.
阅读论文全文,请访问https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-017-0419-x?utm_source=WeChat&utm_medium=Social_media_organic&utm_content=DaiDen-BMC-BMC_Biology-Biology-China&utm_campaign=BMCF_USG_Article
期刊介绍:BMC Biology is an open access journal publishing outstanding research in all areas of biology, with a publication policy that combines selection for broad interest and importance with a commitment to serving authors well.
2016 Journal Metrics:
Citation Impact
6.779 - 2-year Impact Factor
7.556 - 5-year Impact Factor
1.377 - Source Normalized Impact per Paper (SNIP)
(来源:科学网)
特别声明:本文转载仅仅是出于传播信息的需要,并不意味着代表本网站观点或证实其内容的真实性;如其他媒体、网站或个人从本网站转载使用,须保留本网站注明的“来源”,并自负版权等法律责任;作者如果不希望被转载或者联系转载稿费等事宜,请与我们接洽。