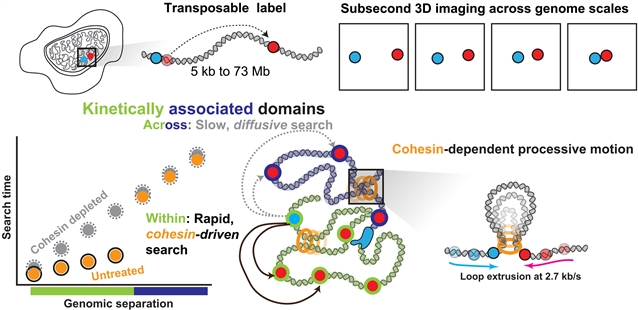
在这里,该课题组研究人员介绍了一种用自定位荧光标记标记哺乳动物染色体并在超分辨率下跟踪它们的方法。该研究团队发现由亚兆基距离分隔的序列在几十秒内转变为接近。这种快速搜索依赖于内聚蛋白,并且只在域内显示。领域边界是搜索过程的动力障碍,而不是结构边界。顺式相互作用的基因组分离依赖的搜索时间尺度违反了对扩散的预测,表明是由马达驱动的折叠。研究组还发现了每秒2.7千碱基的黏结蛋白依赖的进程运动。总之,这些多尺度动力学揭示了基因组在动力学相关域中的组织。
据了解,基因组功能需要调控基因组运动。然而,直接观察体内运动的工具在覆盖范围和分辨率上受到限制。
附:英文原文
Title: Kinetic organization of the genome revealed by ultraresolution multiscale live imaging
Author: Joo Lee, Liang-Fu Chen, Simon Gaudin, Kavvya Gupta, Ana Novacic, Andrew Spakowitz, Alistair Nicol Boettiger
Issue&Volume: 2025-09-18
Abstract: Genome function requires regulated genome motion. However, tools to directly observe this motion in vivo have been limited in coverage and resolution. Here we introduce an approach to tile mammalian chromosomes with self-mapping fluorescent labels and track them at ultraresolution. We find that sequences separated by submegabase distances transition to proximity in tens of seconds. This rapid search is dependent on cohesin and is exhibited only within domains. Domain borders act as kinetic impediments to this search process, rather than structural boundaries. The genomic separation–dependent scaling of the search time for cis interactions violated predictions of diffusion, suggesting motor-driven folding. We also uncover cohesin-dependent processive motion at 2.7 kilobases per second. Together, these multiscale dynamics reveal the organization of the genome into kinetically associated domains.
DOI: adx2202
Source: https://www.science.org/doi/10.1126/science.adx2202
