美国国家癌症研究所Maria Teresa Landi小组的最新研究揭示LINE-1在肺腺癌演变中的作用。2025年12月10日,国际知名学术期刊《自然》发表了这一成果。
在对1024例肺腺癌(LUADs)进行的景观分析中,课题组利用深度全基因组测序和多组学数据,鉴定出542例LUADs具有不同的克隆结构。在这个群体中,该课题组观察到基于吸烟暴露、祖先和性别的不同进化轨迹。来自吸烟者的LUAD在KRAS中显示了大量与烟草相关的C:G>A:T驱动突变和短亚克隆多样化。从不吸烟(以下简称不吸烟者)的LUAD患者显示出与SBS5和SBS40a突变特征相关的拷贝数改变和EGFR突变的早期发生。含有EGFR突变的肿瘤表现出较长的潜伏期,特别是在欧洲血统的女性个体中。来自亚洲从不吸烟者的肿瘤表现出短暂的克隆进化。
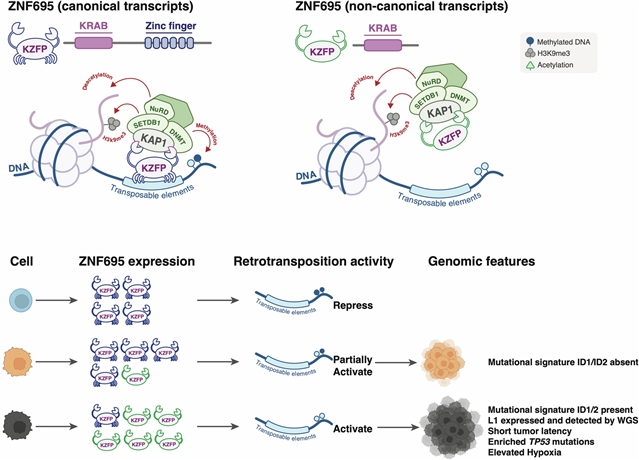
重要的是,该课题组研究人员发现突变特征ID24是以前未被识别的LUAD进化机制的标记。含有ID2的肿瘤潜伏期短,与L1启动子去甲基化相关的长穿插核元件-1 (LINE-1,以下简称L1)反转录转座子活性高。这些肿瘤表现出侵袭性表型,具有基因组不稳定性,缺氧评分升高,新抗原负担低,转移倾向和总生存率差。重新激活的l1逆转录转位诱导的突变可能有助于突变特征ID2,包括通过调控转录因子ZNF695, KZFP家族的一员5。LUAD演变的复杂性为筛查和治疗计划带来了挑战和机遇。
研究人员表示,了解肺癌的进化可以找到阻断其生长的工具。
附:英文原文
Title: Uncovering the role of LINE-1 in the evolution of lung adenocarcinoma
Author: Zhang, Tongwu, Zhao, Wei, Wirth, Christopher, Daz-Gay, Marcos, Yin, Jinhu, Cecati, Monia, Marchegiani, Francesca, Hoang, Phuc H., Leduc, Charles, Baine, Marina K., Travis, William D., Sholl, Lynette M., Joubert, Philippe, Sang, Jian, McElderry, John P., Antony, Michelle, Klein, Alyssa, Khandekar, Azhar, Hartman, Caleb, Rosenbaum, Jennifer, Coln-Matos, Frank J., Miraftab, Mona, Saha, Monjoy, Lee, Olivia W., Jones, Kristine M., Caporaso, Neil E., Wong, Maria Pik, Leung, Kin Chung, Hsiung, Chao Agnes, Chen, Chih-Yi, Edell, Eric S., Santamara, Jacobo Martnez, Schabath, Matthew B., Yendamuri, Sai S., Manczuk, Marta, Lissowska, Jolanta, witkowska, Beata, Mukeria, Anush, Shangina, Oxana, Zaridze, David, Holcatova, Ivana, Mates, Dana, Milosavljevic, Sasa, Savic, Milan, Boss, Yohan, Rothberg, Bonnie E. Gould, Christiani, David C., Gaborieau, Valerie, Brennan, Paul, Liu, Geoffrey, Hofman, Paul, Homer, Robert, Yang, Soo-Ryum, Pesatori, Angela C., Consonni, Dario, Yang, Lixing, Zhu, Bin, Shi, Jianxin, Brown, Kevin, Rothman, Nathaniel, Chanock, Stephen J., Alexandrov, Ludmil B., Choi, Jiyeon, Cardelli, Maurizio, Lan, Qing, Nowak, Martin A.
Issue&Volume: 2025-12-10
Abstract: Understanding lung cancer evolution can identify tools for intercepting its growth1,2. Here, in a landscape analysis of 1,024 lung adenocarcinomas (LUADs) with deep whole-genome sequencing integrated with multiomic data, we identified 542 LUADs with a diverse clonal architecture. In this group, we observed divergent evolutionary trajectories based on tobacco smoking exposure, ancestry and sex. LUAD from smokers showed an abundance of tobacco-related C:G>A:T driver mutations3 in KRAS and short subclonal diversification. LUAD in people who have never smoked (hereafter, never-smokers) showed early occurrence of copy-number alterations and EGFR mutations associated with SBS5 and SBS40a mutational signatures. Tumours containing EGFR mutations exhibited long latency, particularly in female individuals of European-ancestry. Tumours from Asian never-smokers showed a short clonal evolution. Importantly, we found that the mutational signature ID24 is a marker of a previously unrecognized mechanism for LUAD evolution. Tumours with ID2 showed short latency and high long interspersed nuclear element-1 (LINE-1, hereafter L1) retrotransposon activity linked to L1 promoter demethylation. These tumours exhibited an aggressive phenotype with genomic instability, elevated hypoxia scores, low neoantigen burden, metastasis propensity and poor overall survival. Reactivated L1-retrotransposition-induced mutagenesis probably contributes to the mutational signature ID2, including through the regulation of the transcriptional factor ZNF695, a member of the KZFP family5. The complex nature of LUAD evolution creates both challenges and opportunities for screening and treatment plans.
DOI: 10.1038/s41586-025-09825-y
Source: https://www.nature.com/articles/s41586-025-09825-y
Nature:《自然》,创刊于1869年。隶属于施普林格·自然出版集团,最新IF:69.504
官方网址:http://www.nature.com/
投稿链接:http://www.nature.com/authors/submit_manuscript.html
