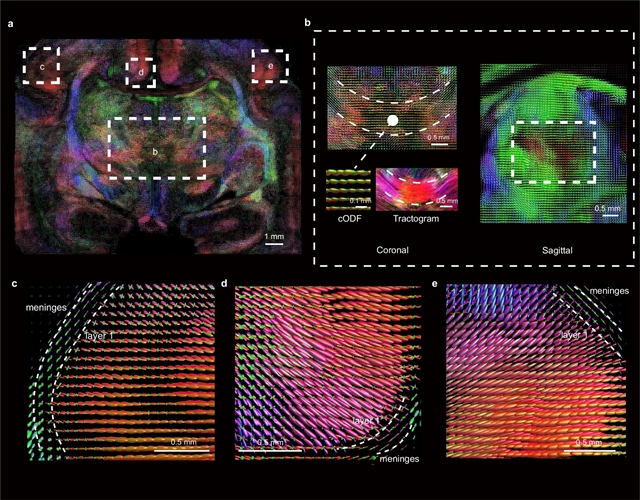
中国科学院深圳先进技术研究院徐放小组的一项最新研究提出了基于细胞结构组织的全脑纤维束重建。这一研究成果于2025年11月3日发表在国际顶尖学术期刊《自然—方法学》上。
利用全脑高通量荧光成像,课题组开发了一种基于细胞结构的链路估计(CABLE)方法,用于在细胞分辨率下精确绘制纤维束。该方法根据核仁或体细胞形状的固有各向异性和相邻细胞的空间排列来推断纤维方向。通过追踪猴子大脑中病毒标记的单个轴突,验证了推断的纤维束。CABLE方法可以解开扩散磁共振成像纤维束成像中不确定的复杂交叉或弯曲纤维束,实现对狨猴和猕猴大脑全脑纤维束的精确重建。最后,课题组人员应用CABLE对新生儿病变脑组织的轴突纤维异常进行快速制图,为人脑纤维束的高分辨率脑制图建立路径。
据了解,轴突轨迹的绘制对于理解大脑组织至关重要。
附:英文原文
Title: Whole-brain reconstruction of fiber tracts based on cytoarchitectonic organization
Author: Zhang, Yue, Song, Tao, Yang, Chao-Yu, Shen, Yan, Yang, Yi, Hu, Xiaowei, Zhao, Rubin, Wang, Weibo, Wang, Xin, Liu, Hean, Yan, Haotian, Jia, Chunyun, Liu, Yang, Wu, Xueyan, Huang, Huizhi, Hu, Xintian, Zhou, Jiang-Ning, Tan, Liming, Lau, Pak-Ming, Wang, Hao, Poo, Mu-Ming, Zhou, Pengcheng, Bi, Guo-Qiang, Xiao, Yanyang, Liu, Cirong, Xu, Fang
Issue&Volume: 2025-11-03
Abstract: Mapping of axon trajectories is crucial for understanding brain organization. Using whole-brain high-throughput fluorescence imaging, we developed a cytoarchitecture-based link estimation (CABLE) method for accurate fiber tract mapping at cellular resolution. This method infers the fiber direction from the inherent anisotropy of the nucleus or soma shape and spatial arrangement of adjacent cells. The inferred fiber tracts were validated by tracing virally labeled individual axons in the monkey brain. This CABLE method could disentangle complex intersecting or bending fibers that were uncertain in diffusion magnetic resonance imaging tractography, allowing accurate brain-wide fiber tract reconstruction in marmoset and macaque brains. Finally, we applied CABLE for rapid mapping of axon fiber abnormalities in diseased neonatal human brain tissues, establishing a path for high-resolution brain mapping of fiber tracts in the human brain.
DOI: 10.1038/s41592-025-02865-2
Source: https://www.nature.com/articles/s41592-025-02865-2
Nature Methods:《自然—方法学》,创刊于2004年。隶属于施普林格·自然出版集团,最新IF:47.99
官方网址:https://www.nature.com/nmeth/
投稿链接:https://mts-nmeth.nature.com/cgi-bin/main.plex
