热力学原理将体外转录因子的亲和力与细胞中的单分子染色质状态联系起来,这一成果由斯坦福大学William J. Greenleaf研究团队经过不懈努力而取得。这一研究成果发表在2025年11月26日出版的国际学术期刊《细胞》上。
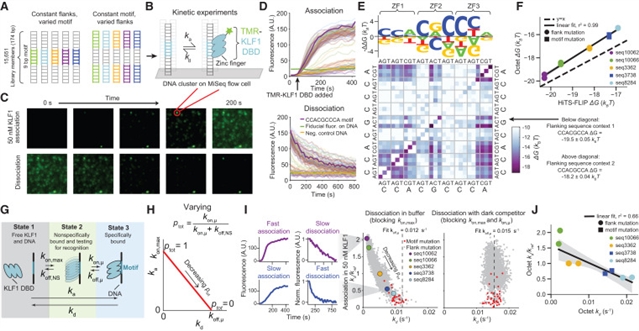
为了解决这些问题,研究人员在高通量下,定量比较了体外TF结合率和亲和力与体内单分子TF和核小体占用率以及体内衍生的深度学习模型。研究小组发现40倍侧翼序列对亲和力的影响与远端侧翼调谐TF搜索参数一致,并被线性能量模型捕获。基序识别概率,而不是在结合状态下的时间,驱动亲和力变化,并且在体外和细胞核测量显示一致的,几分钟长的TF停留时间。最后,体外生物物理参数预测了体内序列偏好和单分子染色质状态的未知基序语法。
研究人员表示,调控转录因子(TF)结合和可接近染色质形成的分子细节尚未定量了解,包括序列上下文如何调节亲和力,TF如何搜索DNA, TF占用动力学以及基序语法如何协调结合。
附:英文原文
Title: Thermodynamic principles link in vitro transcription factor affinities to single-molecule chromatin states in cells
Author: Julia M. Schaepe, Torbjrn Fries, Benjamin R. Doughty, Vivekanandan Ramalingam, Betty B. Liu, Olivia J. Crocker, Georgi K. Marinov, Michaela M. Hinks, Emil Marklund, William J. Greenleaf
Issue&Volume: 2025-11-26
Abstract: The molecular details governing transcription factor (TF) binding and the formation of accessible chromatin are not yet quantitatively understood—including how sequence context modulates affinity, how TFs search DNA, the kinetics of TF occupancy, and how motif grammars coordinate binding. To resolve these questions for a human TF, erythroid Krüppel-like factor (eKLF/KLF1), we quantitatively compare, in high throughput, in vitro TF binding rates and affinities with in vivo single-molecule TF and nucleosome occupancies and in vivo-derived deep learning models. We find that 40-fold flanking sequence effects on affinity are consistent with distal flanks tuning TF search parameters and captured by a linear energy model. Motif recognition probability, rather than time in the bound state, drives affinity changes, and in vitro and in nuclei measurements exhibit consistent, minutes-long TF residence times. Finally, in vitro biophysical parameters predict in vivo sequence preferences and single-molecule chromatin states for unseen motif grammars.
DOI: 10.1016/j.cell.2025.11.008
Source: https://www.cell.com/cell/abstract/S0092-8674(25)01300-5
