威康桑格研究所Iñigo Martincorena团队近日取得一项新成果。经过不懈努力,他们开发出群体尺度上的体细胞突变与选择。这一研究成果于2025年10月8日发表在国际顶尖学术期刊《自然》上。
随着年龄的增长,许多组织被携带体细胞驱动突变的微观克隆定植。这些克隆中的一些代表了迈向癌症的第一步,而另一些则可能导致衰老和其他疾病。
然而,由于在小克隆中检测突变的挑战,他们对这一现象的理解仍然有限。
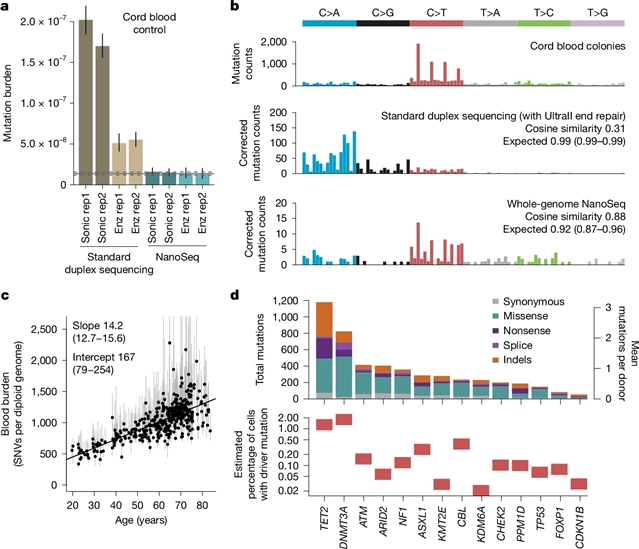
在这里,该课题组研究人员介绍了一种新版本的纳米测序(NanoSeq),这是一种错误率低于每十亿碱基对5个错误的双工测序方法,可兼容全外显子组和靶向捕获。具有单分子灵敏度的多克隆样品深度测序同时描述大量克隆,提供准确的突变率,特征和驱动频率在任何组织。利用靶向NanoSeq对1042份非侵入性口腔上皮样本和371份来自双胞胎队列的血液样本进行分析,研究小组报告了一个极其丰富的选择景观,在口腔上皮中有46个基因处于正选择状态,超过62000个驱动突变和必要基因的负选择证据。许多基因在编码和非编码位点上的高分辨率选择图被获得:一种体内饱和诱变的形式。多变量回归模型使突变流行病学能够研究暴露和癌症风险因素,如年龄、烟草或酒精,如何改变体细胞突变的获得或选择。精确的单分子测序为研究早期癌变、癌症预防以及体细胞突变在衰老和疾病中的作用提供了有力的工具。
附:英文原文
Title: Somatic mutation and selection at population scale
Author: Lawson, Andrew R. J., Abascal, Federico, Nicola, Pantelis A., Lensing, Stefanie V., Roberts, Amy L., Kalantzis, Georgios, Baez-Ortega, Adrian, Brzozowska, Natalia, El-Sayed Moustafa, Julia S., Vaitkute, Dovile, Jakupovic, Belma, Nessa, Ayrun, Wadge, Samuel, sterdahl, Marc F., Paterson, Anna L., Rassl, Doris M., Alcantara, Raul E., ONeill, Laura, Widaa, Sara, Austin-Guest, Siobhan, Neville, Matthew D. C., Przybilla, Moritz J., Cheng, Wei, Morra, Maria, Sykes, Lucy, Mayho, Matthew, Mller-Sienerth, Nicole, Williams, Nicholas, Alexander, Diana, Harvey, Luke M. R., Clarke, Thomas, Byrne, Alex, Blundell, Jamie R., Young, Matthew D., Mahbubani, Krishnaa T. A., Saeb-Parsy, Kourosh, Martin, Hilary C., Stratton, Michael R., Campbell, Peter J., Rahbari, Raheleh, Small, Kerrin S., Martincorena, Iigo
Issue&Volume: 2025-10-08
Abstract: As we age, many tissues become colonized by microscopic clones carrying somatic driver mutations1,2,3,4,5,6,7. Some of these clones represent a first step towards cancer whereas others may contribute to ageing and other diseases. However, our understanding of this phenomenon remains limited due to the challenge of detecting mutations in small clones. Here we introduce a new version of nanorate sequencing (NanoSeq)8, a duplex sequencing method with an error rate lower than five errors per billion base pairs, which is compatible with whole-exome and targeted capture. Deep sequencing of polyclonal samples with single-molecule sensitivity simultaneously profiles large numbers of clones, providing accurate mutation rates, signatures and driver frequencies in any tissue. Applying targeted NanoSeq to 1,042 non-invasive samples of oral epithelium and 371 blood samples from a twin cohort, we report an extremely rich selection landscape, with 46 genes under positive selection in oral epithelium, more than 62,000 driver mutations and evidence of negative selection in essential genes. High-resolution maps of selection across coding and non-coding sites are obtained for many genes: a form of in vivo saturation mutagenesis. Multivariate regression models enable mutational epidemiology studies on how exposures and cancer risk factors, such as age, tobacco or alcohol, alter the acquisition or selection of somatic mutations. Accurate single-molecule sequencing provides a powerful tool to study early carcinogenesis, cancer prevention and the role of somatic mutations in ageing and disease.
DOI: 10.1038/s41586-025-09584-w
Source: https://www.nature.com/articles/s41586-025-09584-w
Nature:《自然》,创刊于1869年。隶属于施普林格·自然出版集团,最新IF:69.504
官方网址:http://www.nature.com/
投稿链接:http://www.nature.com/authors/submit_manuscript.html
