基因组-蛋白质组图谱揭示了自然变异如何驱动蛋白质组多样性和形状适应性,这一成果由
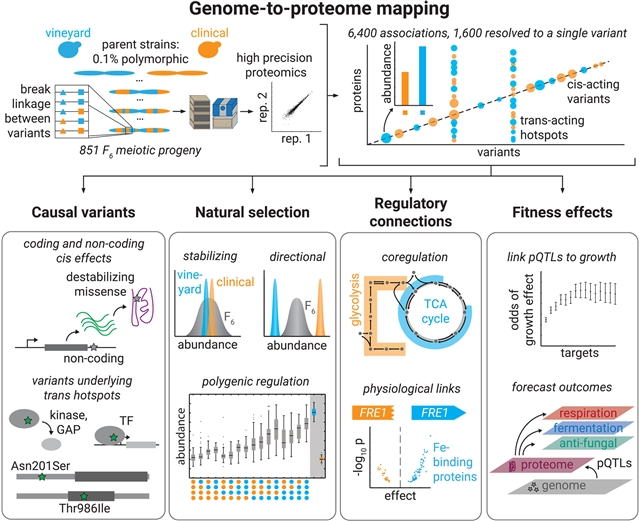
在这项工作中,该课题组通过在适应不同环境的两种酵母菌株之间的800个后代中绘制基因组-蛋白质组关系来解决这一问题。尽管亲本之间的遗传距离不大,但该团队观察到显著的蛋白质组多样性,并绘制了超过6400种基因型-蛋白质关联,其中超过1600种与个体遗传变异有关。蛋白质组适应来自于一个保守的顺式和反式调节变异网络,这些变异通常来自于传统上与基因调节无关的蛋白质。这个图谱使他们能够预测不同条件下的有机体适应性效应。通过以前所未有的分辨率连接基因组和蛋白质组学景观,他们的研究为预测自然遗传变异的表型结果提供了一个框架。
研究人员表示,理解遗传变异如何转化为复杂的表型仍然是一个根本性的挑战。
附:英文原文
Title: A genome-to-proteome map reveals how natural variants drive proteome diversity and shape fitness
Author: Christopher M. Jakobson, Johannes Hartl, Pauline Trébulle, Michael Mülleder, Daniel F. Jarosz, Markus Ralser
Issue&Volume: 2025-10-09
Abstract: Understanding how genetic variation translates into complex phenotypes remains a fundamental challenge. In this work, we address this by mapping genome-to-proteome relationships in 800 progeny of a cross between two yeast strains adapted to distinct environments. Despite the modest genetic distance between the parents, we observed notable proteomic diversity and mapped more than 6400 genotype-protein associations, with more than 1600 linked to individual genetic variants. Proteomic adaptation emerged from a conserved network of cis- and trans-regulatory variants, often originating from proteins not traditionally linked to gene regulation. This atlas allowed us to forecast organismal fitness effects across diverse conditions. By connecting genomic and proteomic landscapes at unprecedented resolution, our study provides a framework for predicting the phenotypic outcomes of natural genetic variation.
DOI: adu3198
Source: https://www.science.org/doi/10.1126/science.adu3198
