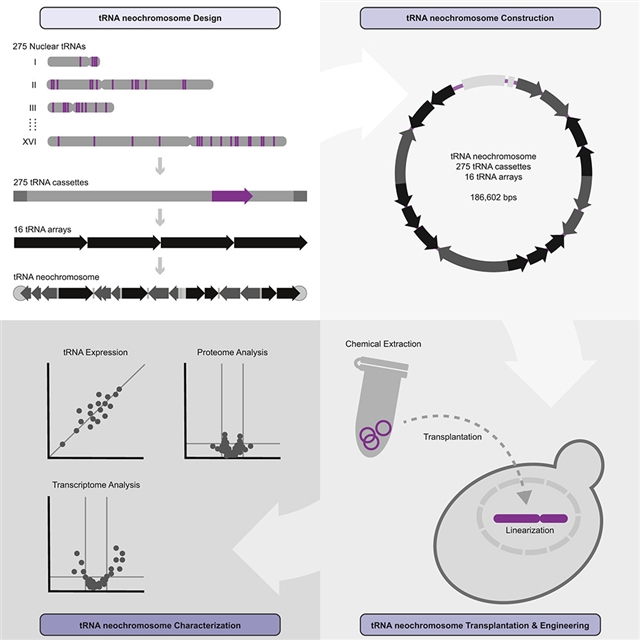
英国曼彻斯特大学蔡毅之研究组报道了酵母中tRNA新染色体的设计、构建和功能表征。相关论文发表在2023年11月8日出版的《细胞》杂志上。
他们报道了tRNA新染色体的设计、构建和表征,这是一种设计染色体,可作为酿酒酵母天然补体的额外的从头配对。为了解决Sc2.0项目的核心设计原则之一,这条约190 kb的tRNA新染色体容纳了所有275个重新定位的核tRNA基因。为了最大限度地提高稳定性,设计结合了来自非S的正交遗传元素。酿酒酵母科的酵母种。283个rox重组位点的存在使正交tRNA SCRaMbLE系统成为可能。在酵母中构建后,他们获得了强有力的选择力的证据,表现为细胞倍性的自发加倍。
此外,研究人员还通过tRNA测序、转录组学、蛋白质组学、核小体作图、复制谱、FISH和Hi-C来研究tRNA新染色体的行为和功能。它的构建证明了酵母模型的显著可追溯性,并为直接测试围绕这些基本非编码RNA的假设提供了机会。
附:英文原文
Title: Design, construction, and functional characterization of a tRNA neochromosome in yeast
Author: Daniel Schindler, Roy S.K. Walker, Shuangying Jiang, Aaron N. Brooks, Yun Wang, Carolin A. Müller, Charlotte Cockram, Yisha Luo, Alicia García, Daniel Schraivogel, Julien Mozziconacci, Noah Pena, Mahdi Assari, María del Carmen Sánchez Olmos, Yu Zhao, Alba Ballerini, Benjamin A. Blount, Jitong Cai, Lois Ogunlana, Wei Liu, Katarina Jnsson, Dariusz Abramczyk, Eva Garcia-Ruiz, Tomasz W. Turowski, Reem Swidah, Tom Ellis, Tao Pan, Francisco Antequera, Yue Shen, Conrad A. Nieduszynski, Romain Koszul, Junbiao Dai, Lars M. Steinmetz, Jef D. Boeke, Yizhi Cai
Issue&Volume: 2023-11-08
Abstract: Here, we report the design, construction, and characterization of a tRNA neochromosome, a designer chromosome that functions as an additional, de novo counterpart to the native complement of Saccharomyces cerevisiae. Intending to address one of the central design principles of the Sc2.0 project, the ~190-kb tRNA neochromosome houses all 275 relocated nuclear tRNA genes. To maximize stability, the design incorporates orthogonal genetic elements from non-S. cerevisiae yeast species. Furthermore, the presence of 283 rox recombination sites enables an orthogonal tRNA SCRaMbLE system. Following construction in yeast, we obtained evidence of a potent selective force, manifesting as a spontaneous doubling in cell ploidy. Furthermore, tRNA sequencing, transcriptomics, proteomics, nucleosome mapping, replication profiling, FISH, and Hi-C were undertaken to investigate questions of tRNA neochromosome behavior and function. Its construction demonstrates the remarkable tractability of the yeast model and opens up opportunities to directly test hypotheses surrounding these essential non-coding RNAs.
DOI: 10.1016/j.cell.2023.10.015
Source: https://www.cell.com/cell/fulltext/S0092-8674(23)01130-3
