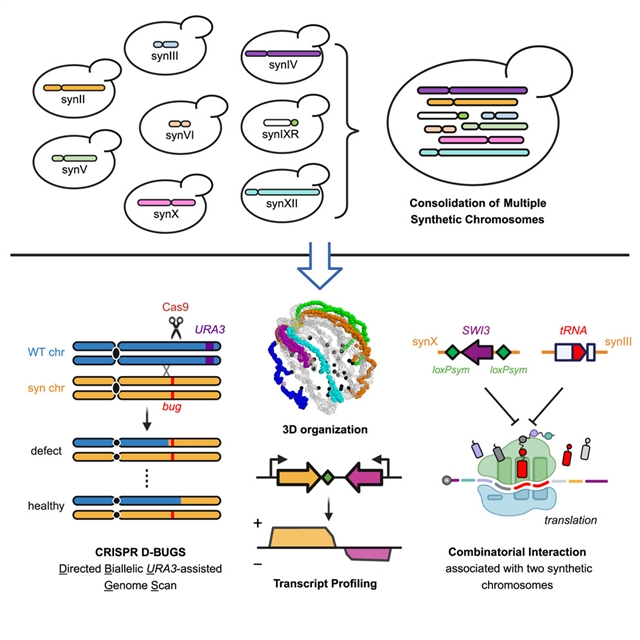
美国纽约大学朗格尼医学院Jef D. Boeke课题组提出了调试和巩固多个合成染色体揭示组合遗传相互作用。这一研究成果发表在2023年11月8日出版的国际学术期刊《细胞》上。
他们描述了多个合成染色体的巩固,使用先进的核内重复交叉与tRNA表达盒产生6.5个合成染色体的菌株。利用Hi-C和长读直接RNA测序对三维染色体组织和转录异构体进行了评估。他们开发了CRISPR定向双等位基因URA3辅助基因组扫描,或“CRISPR D-BUGS”,以绘制由特定设计修饰(称为“bug”)引起的表型变异。
他们首先对合成染色体II (synII)中的一个缺陷进行了精细定位,然后发现了与synII和synX相关的组合相互作用,揭示了转录调控、肌醇代谢和tRNASerCGA丰度之间意想不到的遗传相互作用。最后,为了加快整合,他们采用染色体替换合并最大的染色体(synIV),从而在一个菌株中整合了50%的Sc2.0基因组。
据介绍,Sc2.0项目正在从零开始构建一个真核生物合成基因组。随着所有Sc2.0染色体的组装,一个重要的里程碑已经实现。
附:英文原文
Title: Debugging and consolidating multiple synthetic chromosomes reveals combinatorial genetic interactions
Author: Yu Zhao, Camila Coelho, Amanda L. Hughes, Luciana Lazar-Stefanita, Sandy Yang, Aaron N. Brooks, Roy S.K. Walker, Weimin Zhang, Stephanie Lauer, Cindy Hernandez, Jitong Cai, Leslie A. Mitchell, Neta Agmon, Yue Shen, Joseph Sall, Viola Fanfani, Anavi Jalan, Jordan Rivera, Feng-Xia Liang, Joel S. Bader, Giovanni Stracquadanio, Lars M. Steinmetz, Yizhi Cai, Jef D. Boeke
Issue&Volume: 2023-11-08
Abstract: The Sc2.0 project is building a eukaryotic synthetic genome from scratch. A major milestone has been achieved with all individual Sc2.0 chromosomes assembled. Here, we describe the consolidation of multiple synthetic chromosomes using advanced endoreduplication intercrossing with tRNA expression cassettes to generate a strain with 6.5 synthetic chromosomes. The 3D chromosome organization and transcript isoform profiles were evaluated using Hi-C and long-read direct RNA sequencing. We developed CRISPR Directed Biallelic URA3-assisted Genome Scan, or “CRISPR D-BUGS,” to map phenotypic variants caused by specific designer modifications, known as “bugs.” We first fine-mapped a bug in synthetic chromosome II (synII) and then discovered a combinatorial interaction associated with synIII and synX, revealing an unexpected genetic interaction that links transcriptional regulation, inositol metabolism, and tRNASerCGA abundance. Finally, to expedite consolidation, we employed chromosome substitution to incorporate the largest chromosome (synIV), thereby consolidating >50% of the Sc2.0 genome in one strain.
DOI: 10.1016/j.cell.2023.09.025
Source: https://www.cell.com/cell/fulltext/S0092-8674(23)01079-6
