芬兰赫尔辛基大学Aarno Palotie团队的最新研究表明,FinnGen能够从一个表型良好的孤立种群中提供遗传见解。相关论文于2023年1月18日在线发表于国际学术期刊《自然》。
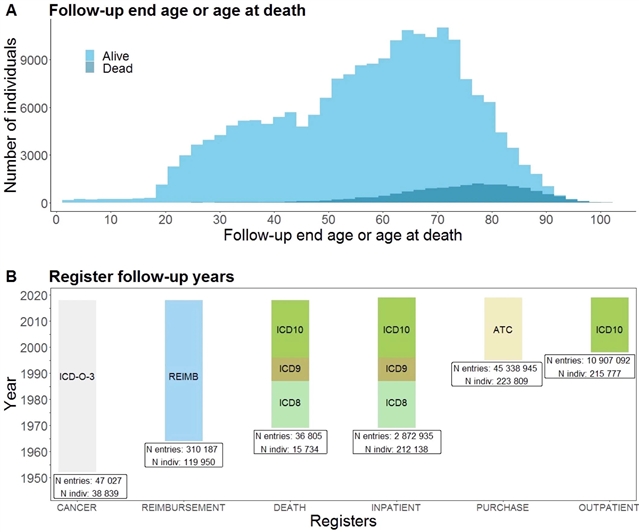
研究人员分析了来自FinnGen的224737名参与者数据,并研究了15种先前在大型全基因组关联研究(GWAS)中研究过的疾病。研究人员还包括来自爱沙尼亚和英国的生物银行数据的元分析。研究人员确定了30个新的关联,主要是低频变异,在芬兰人群中富集。1932种疾病的GWAS也在2496个(771个表型组范围显著性,PWS)独立位点和807个(247个PWS)终点上鉴定了2733个全基因组显著性关联(893个PWS, P<2.6×10-11)。其中,精细映射涉及148(73个PWS)个编码变体,与83(42个PWS)个端点相关。此外,91(47个PWS)在非芬兰的欧洲个体中等位基因频率为<5%,其中62(32个PWS)在芬兰的等位基因频率增加了两倍以上。这些发现证明了瓶颈人群通过低频率、高影响的变异找到常见疾病生物学切入点的力量。
据介绍,像芬兰这样的群体分离物有利于遗传研究,因为缺失等位基因通常集中在少量的低频变异上(0.1%≤小等位基因频率<5%)。这些变体在创建瓶颈中幸存下来,而不是分布在大量的超稀有变体中。尽管这种效应在孟德尔遗传学中得到了很好的证实,但它在常见疾病遗传学中的价值却很少被探索。FinnGen的目标是研究50万芬兰人的基因组和国家健康登记数据。考虑到参与者的中位年龄相对较高(63岁)和医院招聘的很大一部分,fingen在疾病终点方面是丰富的。
附:英文原文
Title: FinnGen provides genetic insights from a well-phenotyped isolated population
Author: Kurki, Mitja I., Karjalainen, Juha, Palta, Priit, Sipil, Timo P., Kristiansson, Kati, Donner, Kati M., Reeve, Mary P., Laivuori, Hannele, Aavikko, Mervi, Kaunisto, Mari A., Loukola, Anu, Lahtela, Elisa, Mattsson, Hannele, Laiho, Pivi, Della Briotta Parolo, Pietro, Lehisto, Arto A., Kanai, Masahiro, Mars, Nina, Rm, Joel, Kiiskinen, Tuomo, Heyne, Henrike O., Veerapen, Kumar, Reger, Sina, Lemmel, Susanna, Zhou, Wei, Ruotsalainen, Sanni, Prn, Kalle, Hiekkalinna, Tero, Koskelainen, Sami, Paajanen, Teemu, Llorens, Vincent, Gracia-Tabuenca, Javier, Siirtola, Harri, Reis, Kadri, Elnahas, Abdelrahman G., Sun, Benjamin, Foley, Christopher N., Aalto-Setl, Katriina, Alasoo, Kaur, Arvas, Mikko, Auro, Kirsi, Biswas, Shameek, Bizaki-Vallaskangas, Argyro, Carpen, Olli, Chen, Chia-Yen, Dada, Oluwaseun A., Ding, Zhihao, Ehm, Margaret G., Eklund, Kari, Frkkil, Martti, Finucane, Hilary, Ganna, Andrea, Ghazal, Awaisa, Graham, Robert R., Green, Eric M., Hakanen, Antti, Hautalahti, Marco, Hedman, sa K., Hiltunen, Mikko
Issue&Volume: 2023-01-18
Abstract: Population isolates such as those in Finland benefit genetic research because deleterious alleles are often concentrated on a small number of low-frequency variants (0.1%≤minor allele frequency<5%). These variants survived the founding bottleneck rather than being distributed over a large number of ultrarare variants. Although this effect is well established in Mendelian genetics, its value in common disease genetics is less explored1,2. FinnGen aims to study the genome and national health register data of 500,000 Finnish individuals. Given the relatively high median age of participants (63years) and the substantial fraction of hospital-based recruitment, FinnGen is enriched for disease end points. Here we analyse data from 224,737 participants from FinnGen and study 15 diseases that have previously been investigated in large genome-wide association studies (GWASs). We also include meta-analyses of biobank data from Estonia and the United Kingdom. We identified 30 new associations, primarily low-frequency variants, enriched in the Finnish population. A GWAS of 1,932 diseases also identified 2,733 genome-wide significant associations (893 phenome-wide significant (PWS), P<2.6×10–11) at 2,496 (771 PWS) independent loci with 807 (247 PWS) end points. Among these, fine-mapping implicated 148 (73 PWS) coding variants associated with 83 (42 PWS) end points. Moreover, 91 (47 PWS) had an allele frequency of <5% in non-Finnish European individuals, of which 62 (32 PWS) were enriched by more than twofold in Finland. These findings demonstrate the power of bottlenecked populations to find entry points into the biology of common diseases through low-frequency, high impact variants.
DOI: 10.1038/s41586-022-05473-8
Source: https://www.nature.com/articles/s41586-022-05473-8
Nature:《自然》,创刊于1869年。隶属于施普林格·自然出版集团,最新IF:69.504
官方网址:http://www.nature.com/
投稿链接:http://www.nature.com/authors/submit_manuscript.html
